Investigate cross sections
It is not in general straight-forward to extract information about applied interaction cross sections and associated mean free path lengths from Geant4. Therefore, a custom mechanism was created in dgcode in the G4XSectDump package, to address this issue. Rather than attempting to piece together the cross sections directly from the various Geant4 data files and C++ algorithms, the mechanism described here instead works by intercepting particles as they are actually being simulated inside an Geant4 job. Not only does this approach have the advantage of reliably extracting the cross sections, it in principle also works with any material and physics list.
Note
If your material is defined by an NCrystal cfg-string,
you can additionally use the nctool command to investigate the material,
to get more detailed information about the thermal (\(<5\mathrm{eV}\))
neutron scattering cross sections (for more information see here).
However, non-neutron, non-scattering, or non-thermal cross sections of the
material in the Geant4 simulations are not provided by nctool, and must
be investigated by using the tools discussed on the present page. Diligent
users might wish to investigate a given material using both approaches.
How to extract cross sections
The extraction can be invoked in two ways:
A generic command-line switch,
-x, exist on all sim-scripts (i.e. runsb_tricorder_sim -xif your script is in a package called “TriCorder”). Using this flag while launching a simulation job will result in cross sections being extracted for all combinations of particle types and materials which are actually encountered in the given job.A command-line utility,
sb_g4xsectdump_query, exists for extracting and displaying information for a requested combination of material, particle type and physics list. By default, this command will also launch cross section plots automatically.
More specifically, the results are extracted in the form of custom data files,
each file containing cross section information for one particular combination of
particle type, Geant4 material and Geant4 physics list. In the
XSectParse package are utilities which can be used to parse and analyse
those files, namely the sb_xsectparse_plotfile command and the two Python
modules XSectParse.ParseXSectFile and XSectParse.PlotXSectFile.
The query script
The query script is used to conveniently handle both the launch of Geant4 and the creation and analysis of cross section files, all via a simple interface:
$> sb_g4xsectdump_query -h
Usage: sb_g4xsectdump_query [options]
This script allows you to extract cross sections from Geant4, for given
specified combinations of material, physics list and particle type. Note that
power users might want to fine-tune the granularity of the extraction by
setting environment variables G4XSECTSPY_LOGDELTAE (default value 0.005) and
G4XSECTSPY_NSAMPLE (default value 50).
Options:
-h, --help show this help message and exit
-p PARTICLE, --particle=PARTICLE
Name or PDG code of particle
-l PHYSLIST, --physlist=PHYSLIST
Name of physics list (default QGSP_BIC_HP_EMZ)
-m MATNAME, --material=MATNAME
Material given by NamedMaterialProvider syntax
-s, --noshow Don't show extracted cross sections in interactive
plots
-f, --nofile Don't save plots of extracted cross sections in PDF
files
-w, --wavelengths Show plots versus neutron wavelength rather than
energy
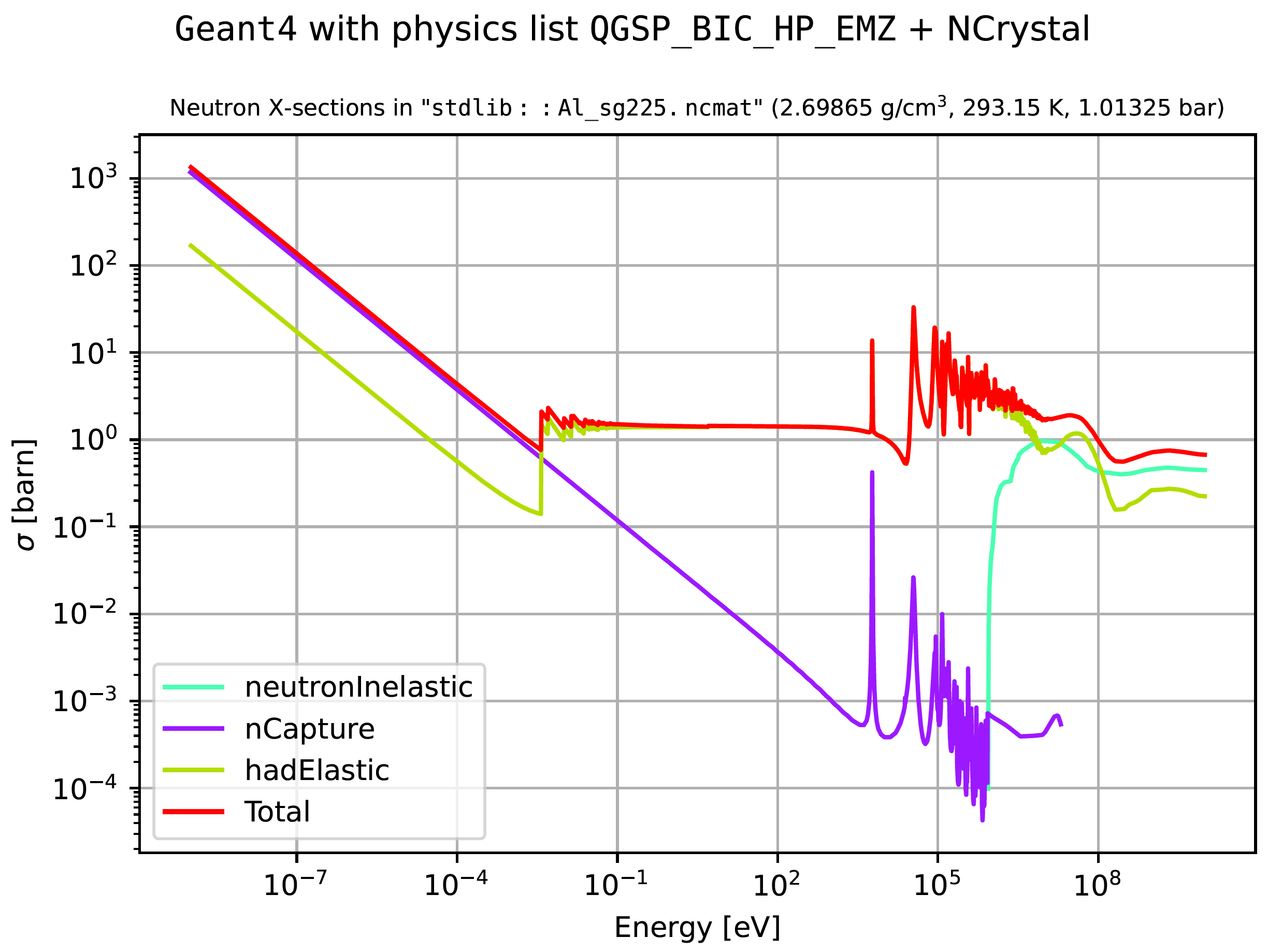
Here is an example where neutron cross sections in aluminium are extracted (refer to the relevant documentation to learn more about the strings defining materials or physics lists):
$> sb_g4xsectdump_query -m 'stdlib::Al_sg225.ncmat' -l QGSP_BIC_HP_EMZ -p neutron
As in fact, QGSP_BIC_HP_EMZ is the default physics list
and neutron is the default particle, we could have simply written:
$> sb_g4xsectdump_query -m 'stdlib::Al_sg225.ncmat'
Note how the material definition string 'stdlib::Al_sg225.ncmat' was quoted
with ' characters. This is in general a good idea, since such strings might
contain special characters which could be otherwise be interpreted by your shell
rather than being passed to sb_g4xsectdump_query.
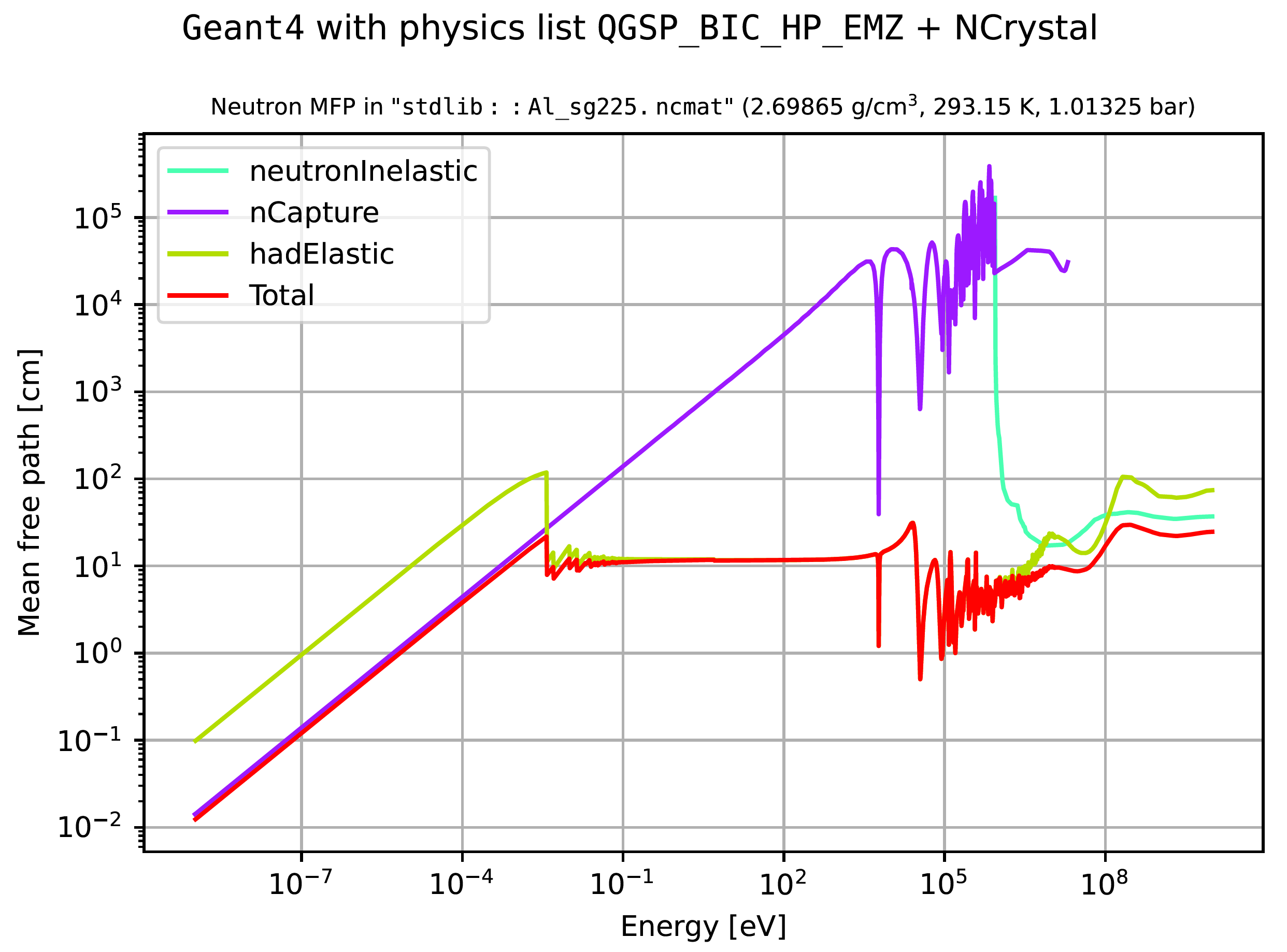
In addition to launching interactive Matplotlib-based cross section plots, either of the above commands produce a list of data files with cross section information, a file with a dump of the G4Material, and files with plots like the following:
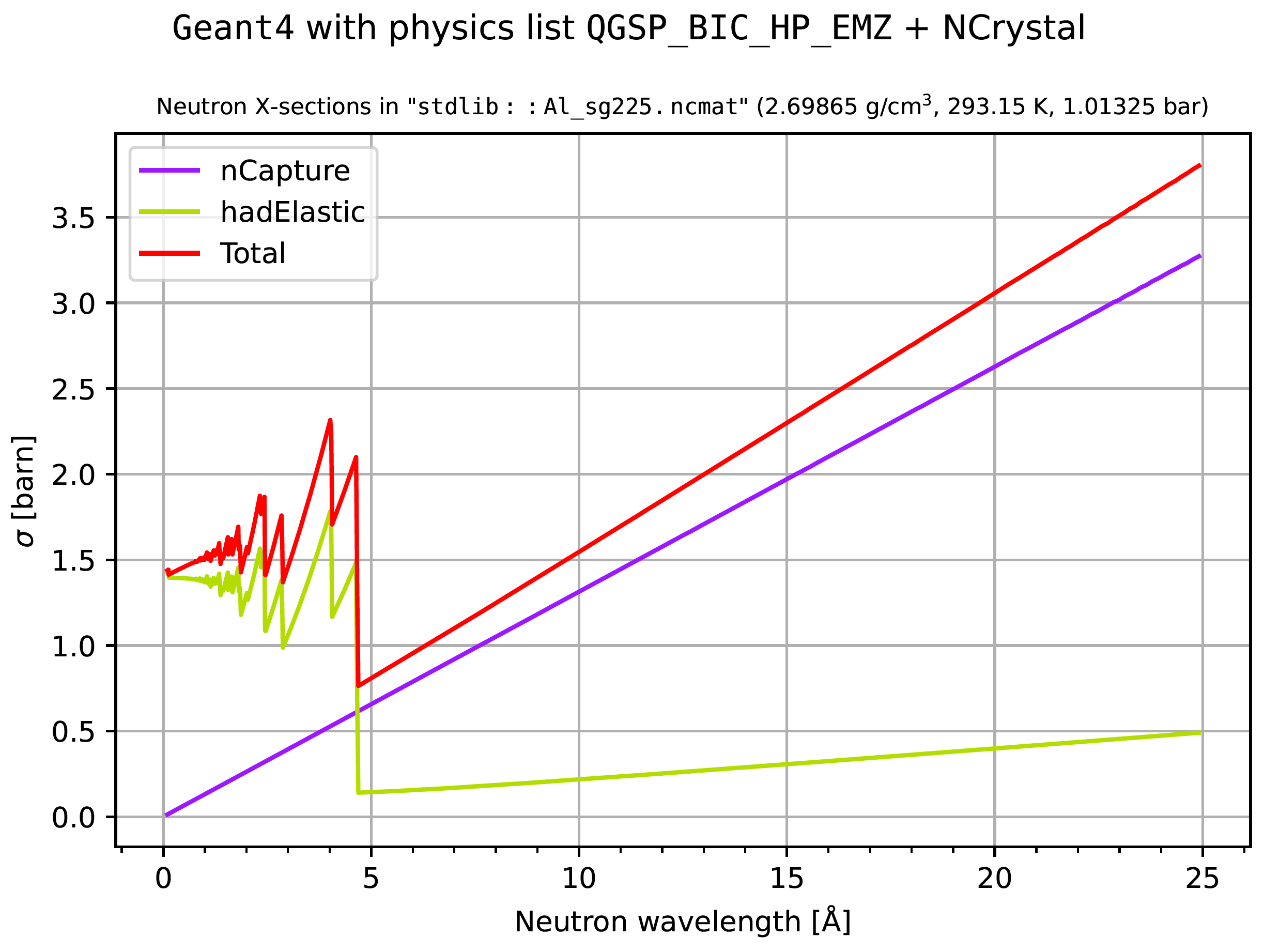
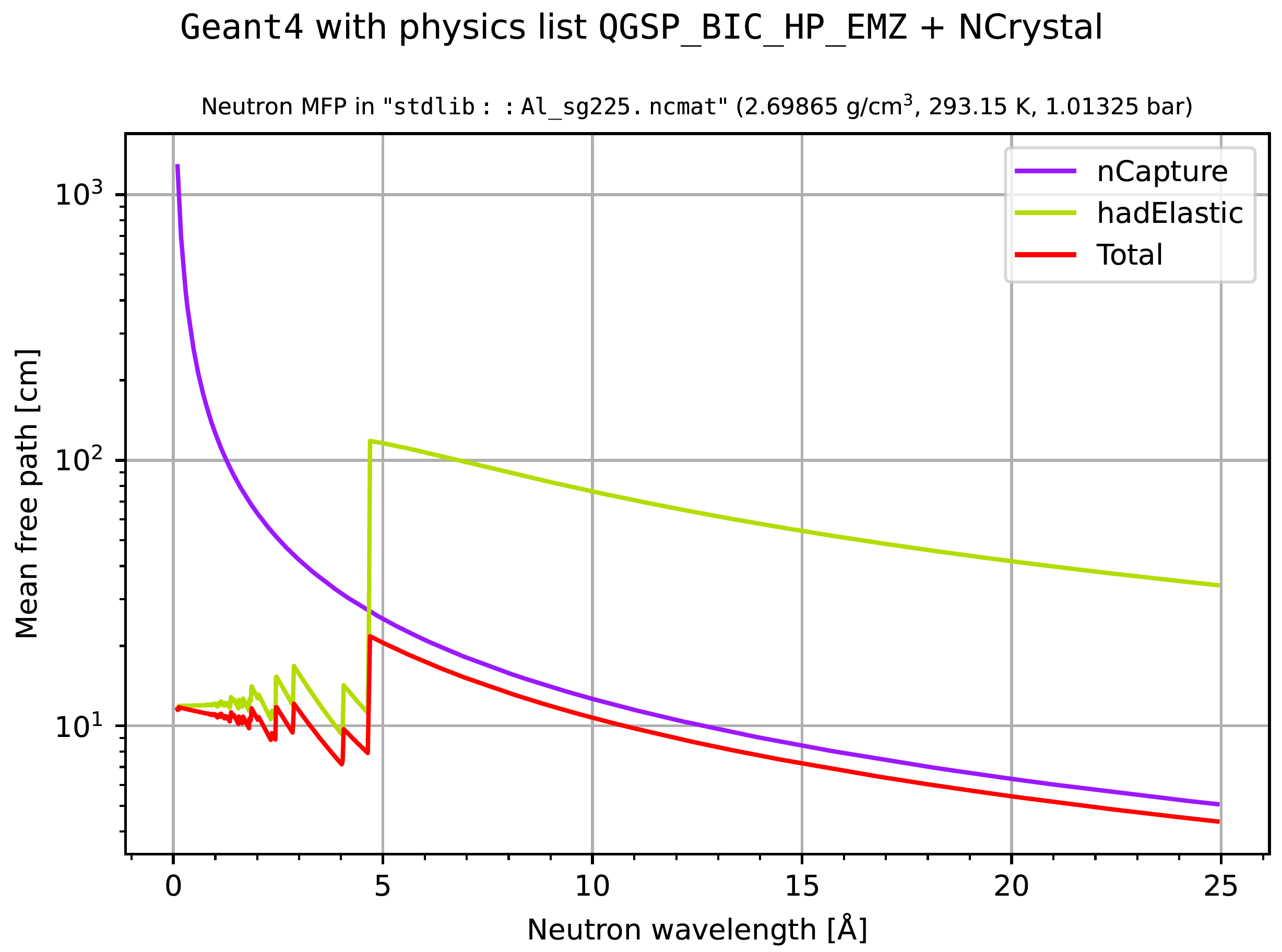
If additionally --wavelengths or -w is supplied on the command line, the
plots would instead show cross sections and mean-free-path lengths as a function
of neutron wavelength (for 0.1 to 25 Å):
To investigate per-atom cross sections of particular elements or isotopes, one can (ab)use the gas-mixture features of NCrystal, to create a simple material which is modelled as a “noblegas” of such atoms. Here is for example how to extract cross sections for Fe56:
sb_g4xsectdump_query -m 'gasmix::Fe56'
This models the material as a gas of monoatomic non-interacting molecules (a
fake noble gas), and at default pressure and temperature values of 1atm and
293.15K respectively. Pressure and temperature could of course be overridden via
additional parameters, but it most likely does not make much sense to worry
about such details anyway, since Fe56 does not actually exist as a gas of
monoatomic molecules. Do note that mean-free-path and macroscopic cross sections
are therefore not really sensible for academic materials like gasmix::Fe56,
but the per-atom absorption and epithermal scattering cross sections are very
much well defined.
Improving precision
As indicated in the usage information dumped above, advanced-users who are worried about random fluctuations or missing narrow peaks due to too low sampling granularity can modify these via environment variables, for instance like this:
$> export G4XSECTSPY_NSAMPLE=100 # reduce fluctuations, default is 50
$> export G4XSECTSPY_LOGDELTAE=0.0001 # increase granularity, default is 0.005
$> sb_g4xsectdump_query -m"gasmix::Fe56"
In case you are wondering why you in the first place would ever need to sample a cross section value at a given energy more than once, it is because some processes in Geant4 actually employ random number generators in the calculation of cross sections, supposedly in an effort to improve various modelling characteristics when averaged over a large number of interactions. It might be worth mentioning here that NCrystal does not employ such schemes.
Parsing utilities
In addition to the query script mentioned above, there are also a few utilities in the XSectParse package for parsing and plotting the contents of cross section files:
The command
sb_xsectparse_plotfilewhich can be run on a cross section file to produce plots of the contents:$> sb_xsectparse_plotfile -h Usage: sb_xsectparse_plotfile [-h|--help] [-l] [xsectfile1] [xsectfile2] [...] Script for quickly plotting the contents of xsectfiles dumped by the G4XSectDump.XSectSpy module (usually through the -x option to a simulation script) -h/--help : Show usage information -l : Display mean free path length rather than cross sections
Python modules for parsing or plotting the contents of the cross section files:
import XSectParse.ParseXSectFile import XSectParse.PlotXSectFile
Implementation and limitations
There are many different kinds of processes in Geant4. Most are discrete, in the sense that they might or might not take place (according to a mean-free-path length based randomisation). These are the only kinds of cross sections extracted by the mechanism discussed on the present page. Other non-discrete processes such as decay and steady energy loss due to ionisation are ignored. For the details, refer to the implementation in XSectSpySteppingAction.hh.