Using MCPL from the command line
The MCPL distribution includes a handy command-line tool, mcpltool, which can be used to either inspect MCPL files, or to carry out a limited number of operations on them. Since release 1.2, it also includes a second tool, pymcpltool, which provides additional inspection features in the form of plots and statistics display.
This page includes a few examples of how the command-line tools can be used, but users are referred to the MCPL paper (section 2.3) for more information.
Examples
A few examples of how to use the command-line tools are provided here. Note that a small sample MCPL file is available at examples/example.mcpl, in case new users would like something to try the mcpltool on.
Inspect file contents
Simply invoking mcpltool or pymcpltool on a file with no additional arguments, results in a summary of the header information being printed, in addition to the particle state information of the first ten particles in the file:
$ mcpltool example.mcpl
Opened MCPL file example.mcpl:
Basic info
Format : MCPL-3
No. of particles : 1006
Header storage : 140 bytes
Data storage : 36216 bytes
Custom meta data
Source : "G4MCPLWriter [G4MCPLWriter]"
Number of comments : 1
-> comment 0 : "Transmission spectrum from 10GeV proton beam on 20cm lead"
Number of blobs : 0
Particle data format
User flags : no
Polarisation info : no
Fixed part. type : no
Fixed part. weight : no
FP precision : single
Endianness : little
Storage : 36 bytes/particle
index pdgcode ekin[MeV] x[cm] y[cm] z[cm] ux uy uz time[ms] weight
0 22 1.2635 -3.5852 -0.81223 20 -0.41453 -0.022799 0.90975 7.3389e-07 1
1 22 2.6273 0.85935 10.196 20 0.0031473 0.75937 0.65065 8.6567e-07 1
2 211 1050.4 2.9741 -0.32269 20 0.24133 0.0099724 0.97039 7.1473e-07 1
3 2112 0.26395 -2.7828 -4.709 20 -0.7575 0.041025 0.65154 2.7656e-05 1
4 2112 0.34922 0.42959 -11.636 20 0.22266 -0.82642 0.51716 1.9382e-05 1
5 2112 1.4445 3.8808 14.263 20 -0.036128 0.47899 0.87708 1.9604e-05 1
6 2112 0.21436 -20.706 0.071227 20 -0.42916 0.43638 0.79082 1.1434e-05 1
7 2112 0.27496 -6.9939 7.9537 20 -0.47614 0.36919 0.79812 6.8358e-05 1
8 2112 0.41955 -3.0206 0.11889 20 -0.62614 0.040539 0.77866 2.7557e-05 1
9 2112 0.64336 -11.788 12.976 20 -0.77018 -0.35919 0.52707 6.1839e-05 1
The -l (limit) and -s (skip) flags can be used to change which particles are printed (use -l0 to print all particles), and -j can be used to suppress the header information.
Extract some particles from a file
Using the --extract flag to mcpltool, it is possible to extract a subset of particles from a file, into a new file. Using the -p flag, one can select according to particle type (2112=neutron, 22=gamma, etc.).
mcpltool --extract -p2112 example.mcpl justneutrons.mcpl
MCPL: Attempting to compress file justneutrons.mcpl with gzip
MCPL: Succesfully compressed file into justneutrons.mcpl.gz
MCPL: Succesfully extracted 726 / 1006 particles from examples/example.mcpl into justneutrons.mcpl.gz
Note that the output file is currently always compressed into .mcpl.gz when possible (this behaviour might change in the future).
You can also use the -l and -s flags to extract particles according to their position in the file, which might for instance be useful to extract a specific interesting particle from a huge file. Here we extract 1 particle starting from position 123:
mcpltool --extract -l1 -s123 examples/example.mcpl selected.mcpl
MCPL: Attempting to compress file selected.mcpl with gzip
MCPL: Succesfully compressed file into selected.mcpl.gz
MCPL: Succesfully extracted 1 / 1006 particles from examples/example.mcpl into selected.mcpl.gz
Merging compatible files
Using the --merge flag to mcpltool, it is possible to merge contents from a list of compatible files into a single new one. Here four existing files are merged, creating newfile.mcpl as a result:
mcpltool --merge newfile.mcpl file1.mcpl file2.mcpl file3.mcpl file4.mcpl
Note that files are considered compatible if and only if they have similar settings and meta-data such as comments and binary blobs. Thus, the resulting file will have the same such settings and meta-data as the originals.
Merging incompatible files (“forcemerge”)
Occasionally, the compatiblity requirements for the –merge option can not be
satisfied. At the expense of discarding descriptive meta-data such as comments and binary
blobs, a brute-force merge of the files can be instead performed with the
--forcemerge flag:
mcpltool --forcemerge newfile.mcpl file1.mcpl file2.mcpl file3.mcpl file4.mcpl
The settings and per-particle storage requirements of the resulting file will automatically be adapted to be such that particles from all the input files can be safely represented. For example, even if just one input file use double-precision storage for its particles, the merged output file will use double-precision for all of its particles.
Due to the loss of meta-data, the usage of the --forcemerge option should be
considered as a last-resort only and is in general not recommended.
Get full usage instructions
Full usage instructions are obtainable with the --help flag:
$ mcpltool --help
Tool for inspecting or modifying Monte Carlo Particle List (.mcpl) files.
The default behaviour is to display the contents of the FILE in human readable
format (see Dump Options below for how to modify what is displayed).
This installation supports direct reading of gzipped files (.mcpl.gz).
Usage:
mcpltool [dump-options] FILE
mcpltool --merge [merge-options] FILE1 FILE2
mcpltool --extract [extract-options] FILE1 FILE2
mcpltool --repair FILE
mcpltool --version
mcpltool --help
Dump options:
By default include the info in the FILE header plus the first ten contained
particles. Modify with the following options:
-j, --justhead : Dump just header info and no particle info.
-n, --nohead : Dump just particle info and no header info.
-lN : Dump up to N particles from the file (default 10). You
can specify -l0 to disable this limit.
-sN : Skip past the first N particles in the file (default 0).
-bKEY : Dump binary blob stored under KEY to standard output.
Merge options:
-m, --merge FILEOUT FILE1 FILE2 ... FILEN
Creates new FILEOUT with combined particle contents from
specified list of N existing and compatible files.
-m, --merge --inplace FILE1 FILE2 ... FILEN
Appends the particle contents in FILE2 ... FILEN into
FILE1. Note that this action modifies FILE1!
--forcemerge [--keepuserflags] FILEOUT FILE1 FILE2 ... FILEN
Like --merge but works with incompatible files as well, at the
heavy price of discarding most metadata like comments and blobs.
Userflags will be discarded unless --keepuserflags is specified.
Extract options:
-e, --extract FILE1 FILE2
Extracts particles from FILE1 into a new FILE2.
-lN, -sN : Select range of particles in FILE1 (as above).
-pPDGCODE : select particles of type given by PDGCODE.
Other options:
-r, --repair FILE
Attempt to repair FILE which was not properly closed, by up-
dating the file header with the correct number of particles.
-t, --text MCPLFILE OUTFILE
Read particle contents of MCPLFILE and write into OUTFILE
using a simple ASCII-based format.
-v, --version : Display version of MCPL installation.
-h, --help : Display this usage information (ignores all other options).
Running instead pymcpltool --help shows identical options as for the
mcpltool, with the exception that the merge, extract and repair options are
absent and that instead options to extract statistics are available:
Stat options:
--stats FILE : Print statistics summary of particle state data from FILE.
--stats --pdf FILE
: Produce PDF file mcpl.pdf with histograms of particle state
data from FILE.
--stats --gui FILE
: Like --pdf, but opens interactive histogram views directly.
Extract statistics from a file
Using the pymcpltool with the --stats flag, it is possible to analyse a file
to get statistics of the contained particles and the distribution of their state
parameters:
pymcpltool --stats example.mcpl
------------------------------------------------------------------------------
nparticles : 1006
sum(weights) : 1006
------------------------------------------------------------------------------
: mean rms min max
------------------------------------------------------------------------------
ekin [MeV] : 52.165 548.838 0.00139345 9724.96
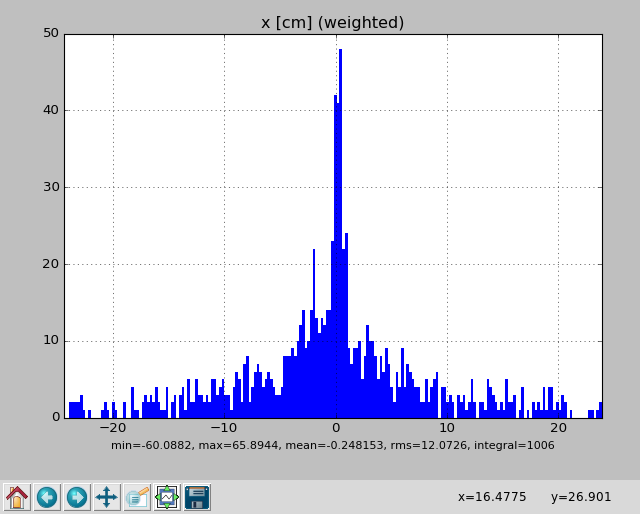
x [cm] : -0.248153 12.0726 -60.0882 65.8944
y [cm] : 2.54021 12.7636 -61.3354 57.6935
z [cm] : 20 0 20 20
ux : 0.0216263 0.487791 -0.994109 0.995773
uy : -0.00868233 0.47584 -0.994904 0.979886
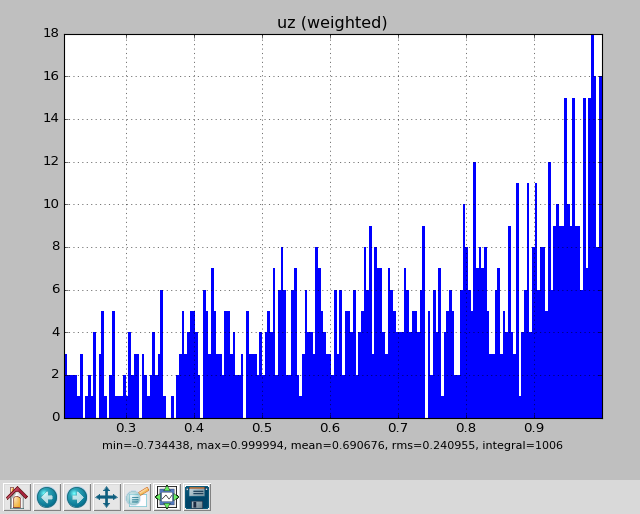
uz : 0.690676 0.240955 -0.734438 0.999994
time [ms] : 5.04828e-05 0.00010785 7.01658e-07 0.00166018
weight : 1 0 1 1
polx : 0 0 0 0
poly : 0 0 0 0
polz : 0 0 0 0
------------------------------------------------------------------------------
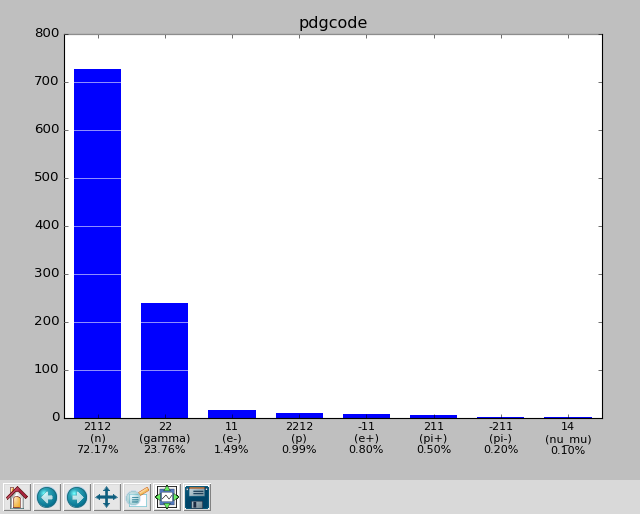
pdgcode : 2112 (n) 726 (72.17%)
22 (gamma) 239 (23.76%)
11 (e-) 15 ( 1.49%)
2212 (p) 10 ( 0.99%)
-11 (e+) 8 ( 0.80%)
211 (pi+) 5 ( 0.50%)
-211 (pi-) 2 ( 0.20%)
14 (nu_mu) 1 ( 0.10%)
[ values ] [ weighted counts ]
------------------------------------------------------------------------------
userflags : 0 (0x00000000) 1006 (100.00%)
[ values ] [ weighted counts ]
------------------------------------------------------------------------------
Or, one can view the parameter distributions graphically by adding --gui:
pymcpltool --stats --gui example.mcpl
Resulting in plots of the file contents like these:
Or one can produce a PDF file like this one
containing the plots by adding instead the --pdf flag:
pymcpltool --stats --pdf example.mcpl
Extract file contents to text file
Using the mcpltool or pymcpltool with the --text flag (available since
MCPL version 1.2.0), it is possible to extract particle data from MCPL files
into simple text (ASCII) files. This might be useful for compatibility with
software expecting data in column-based text files, but the resulting files are
obviously significantly larger and less efficient to use than the original MCPL
files:
mcpltool --text example.mcpl out.txt